Biological systems rely on complex interaction networks at various scales from molecules to organisms. Working in collaboration with experimentalists and through joint PhDs with wet labs, we are building models to understand emergent phenomena in biology as well as predict their temporal evolution. All our projects rely on nonlinear dynamics, stochastic modelling and statistical learning methods.
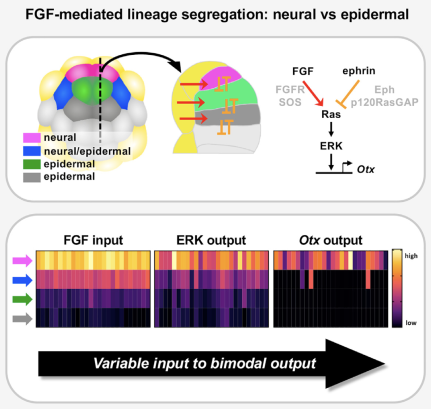
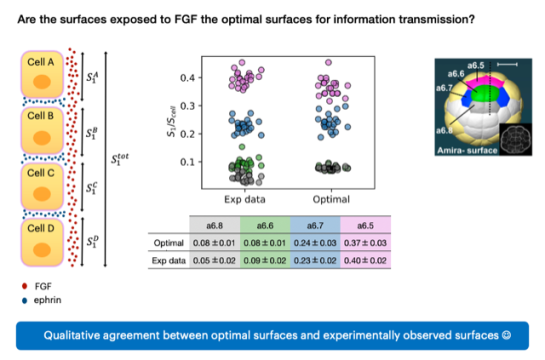
- Embryonic development: How do cells take decisions about their fates? What are the molecular mechanisms underlying these decisions? To answer those questions and understand reproducibility in the process, we focus on the early embryonic development of ascidians which are marine animals with a very simple embryogenesis. We are particularly interested in the link between the geometry of the embryo, via the surfaces of contact between cells, and cell fate assignment. More recently, we started working on information transmission and ask if the geometry of the embryo is optimally set for the establishment of cell fates. Eventually we aim at integrating signaling and mechanics to link the cell fate map to the fertilized egg. This work is done in collaboration with the lab of H. Yasuo, Geneviève Dupont and Aleksandra Walczak.
- Engineering genetic circuits for synthetic biology: In the long term, we aim to design bottom-up microbial cell factories for the production of bioplastics, in collaboration with Eveline Peeters and Wim Vranken. In the short term, we aim to 1) develop and optimise metabolite-responsive biosensor modules and 2) rationally design synthetic regulatory circuits to improve the performance of production pathways in E. coli - the most studied prokaryotic organism.
- Dynamical modeling of enterohepatic circulation: We aim to investigate the progression of metabolic dysfunction-associated steatotic liver disease (MASLD). We are developing a dynamical model of the human enterohepatic circulation to predict bile acid profiles in compartments such as the liver, intestines and portal blood that are difficult to access clinically. We will validate these predictions with clinical data in collaboration with Isabelle Leclercq and Laure-Alix Clerbaux. We are particularly interested in using the model to investigate the role of gut microbiota metabolism in MASLD progression and to test the impact of potential therapeutic interventions.
Papers
Rossana Bettoni, Genevieve Dupont, Aleksandra M. Walczak & Sophie de Buyl
Géraldine Williaume, Sophie de Buyl, Cathy Sirour, Nicolas Haupaix, Kaoru Imai, Yutaka Satou, Geneviève Dupont, Rossana Bettoni, Clare Hudson & Hitoyoshi Yasuo